Downloads
Download


This work is licensed under a Creative Commons Attribution 4.0 International License.
Review
Application of LLMs/Transformer-Based Models for Metabolite Annotation in Metabolomics
Yijiang Liu 1,†, Feifan Zhang 2,†, Yifei Ge 2, Qiao Liu 3, Siyu He 4, and Xiaotao Shen 1,2,5,*
1 School of Chemistry, Chemical Engineering and Biotechnology, Nanyang Technological University,
Singapore 637459, Singapore
2 Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore 308232, Singapore
3 Department of Statistics, Stanford University School of Medicine, Palo Alto, CA 94304, USA
4 Department of Biomedical Data Science, Stanford University School of Medicine, Palo Alto, CA 94304, USA
5 Singapore Phenome Center, Nanyang Technological University, Singapore 636921, Singapore
* Correspondence: xiaotao.shen@ntu.edu.sg
† These authors contributed equally to this work.
Received: 20 December 2024; Revised: 6 January 2025; Accepted: 3 March 2025; Published: 15 April 2025
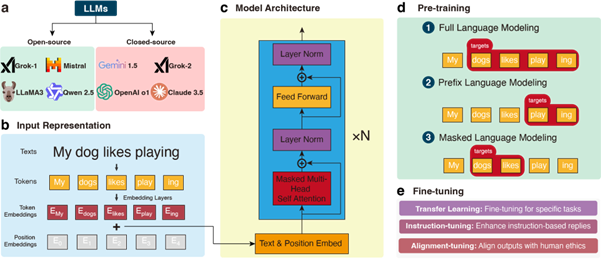
Abstract: Liquid Chromatography-Mass Spectrometry (LC-MS) untargeted metabolomics has become a cornerstone of modern biomedical research, enabling the analysis of complex metabolite profiles in biological systems. However, metabolite annotation, a key step in LC-MS untargeted metabolomics, remains a major challenge due to the limited coverage of existing reference libraries and the vast diversity of natural metabolites. Recent advancements in large language models (LLMs) powered by Transformer architecture have shown significant promise in addressing challenges in data-intensive fields, including metabolomics. LLMs, which when fine-tuned with domain-specific datasets such as mass spectrometry (MS) spectra and chemical property databases, together with other Transformer-based models, excel at capturing complex relationships and processing large-scale data and significantly enhance metabolite annotation. Various metabolomics tasks include retention time prediction, chemical property prediction, and theoretical MS2 spectra generation. For example, methods such as LipiDetective and MS2Mol have shown the potential of machine learning in lipid species prediction and de novo molecular structure annotation directly from MS2 spectra. These tools leverage transformer principles and their integration with LLM frameworks could further expand their utility in metabolomics. Moreover, the ability of LLMs to integrate multi-modal datasets—spanning genomics, transcriptomics, and metabolomics—positions them as powerful tools for systems-level biological analysis. This review highlights the application and future perspectives of Transformer-based LLMs for metabolite annotation of LC-MS metabolomics incorporating with multiomics. Such transformative potential paves the way for enhanced annotation accuracy, expanded metabolite coverage, and deeper insights into metabolic processes, ultimately driving advancements in precision medicine and systems biology.
Keywords:
large language models LC-MS metabolite annotationReferences
- Klassen, A.; Faccio, A.T.; Canuto GA, B.; da Cruz PL, R.; Ribeiro, H.C.; Tavares MF, M.; Sussulini, A. Metabolomics: Definitions and Significance in Systems Biology. In Metabolomics: From Fundamentals to Clinical Applications; Sussulini, A., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 3–17. https://doi.org/10.1007/978-3-319-47656-8_1.
- Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007, 387, 525–527. doi: 10.1007/s00216-006-0687-8
- Emwas, A.-H.M. The Strengths and Weaknesses of NMR Spectroscopy and Mass Spectrometry with Particular Focus on Metabolomics Research. In Metabonomics: Methods and Protocols; Bjerrum, J.T., Ed.; Springer: New York, NY, USA, 2015; pp. 161–193. https://doi.org/10.1007/978-1-4939-2377-9_13.
- Glish, G.L.; Vachet, R.W. The basics of mass spectrometry in the twenty-first century. Nat. Rev. Drug Discov. 2003, 2, 140–150. doi: 10.1038/nrd1011
- Son, A.; Kim, W.; Park, J.; Park, Y.; Lee, W.; Lee, S.; Kim, H. Mass Spectrometry Advancements and Applications for Biomarker Discovery, Diagnostic Innovations, and Personalized Medicine. Int. J. Mol. Sci. 2024, 25, 9880. doi: 10.3390/ijms25189880
- Qin, X.; Hakenjos, J.M.; Li, F. LC-MS-Based Metabolomics in the Identification of Biomarkers Pertaining to Drug Toxicity: A New Narrative. In Biomarkers in Toxicology; Patel, V.B., Preedy, V.R., Rajendram, R., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 1–25. https://doi.org/10.1007/978-3-030-87225-0_34-1.
- Shen, X.; Yan, H.; Wang, C.; Gao, P.; Johnson, C.H.; Snyder, M.P. TidyMass an object-oriented reproducible analysis framework for LC–MS data. Nat. Commun. 2022, 13, 4365. doi: 10.1038/s41467-022-32155-w
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Anal. Biotechnol. 2019, 55, 44–50. doi: 10.1016/j.copbio.2018.07.010
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837.
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. doi: 10.1038/s41592-019-0344-8
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 3. doi: 10.1186/s13321-016-0115-9
- Shen, X.; Wang, R.; Xiong, X.; Yin, Y.; Cai, Y.; Ma, Z.; Liu, N.; Zhu, Z.-J. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat. Commun. 2019, 10, 1516. doi: 10.1038/s41467-019-09550-x
- Shen, X.; Wu, S.; Liang, L.; Chen, S.; Contrepois, K.; Zhu, Z.J.; Snyder, M. metID: An R package for automatable compound annotation for LC−MS-based data. Bioinformatics 2022, 38, 568–569. doi: 10.1093/bioinformatics/btab583
- Shen, X.; Wang, C.; Snyder, M.P. massDatabase: Utilities for the operation of the public compound and pathway database. Bioinformatics 2022, 38, 4650–4651. doi: 10.1093/bioinformatics/btac546
- Nguyen, Q.-H.; Nguyen, H.; Oh, E.C.; Nguyen, T. Current approaches and outstanding challenges of functional annotation of metabolites: A comprehensive review. Brief. Bioinform. 2024, 25, bbae498. doi: 10.1093/bib/bbae498
- Guo, J.; Yu, H.; Xing, S.; Huan, T. Addressing big data challenges in mass spectrometry-based metabolomics. Chem. Commun. 2022, 58, 9979–9990. doi: 10.1039/D2CC03598G
- Huang, K.; Mo, F.; Zhang, X.; Li, H.; Li, Y.; Zhang, Y.; Yi, W.; Mao, Y.; Liu, J.; Xu, Y.; et al. A Survey on Large Language Models with Multilingualism: Recent Advances and New Frontiers. arXiv 2024, arXiv:2405.10936.
- Bharathi Mohan, G.; Prasanna Kumar, R.; Vishal Krishh, P.; Keerthinathan, A.; Lavanya, G.; Meghana, M.K.U.; Sulthana, S. An analysis of large language models: Their impact and potential applications. Knowl. Inf. Syst. 2024, 66, 5047–5070. doi: 10.1007/s10115-024-02120-8
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2023.
- Xu, B.; Poo, M. Large language models and brain-inspired general intelligence. Natl. Sci. Rev. 2023, 10, nwad267. doi: 10.1093/nsr/nwad267
- Zhang, S.; Fan, R.; Liu, Y.; Chen, S.; Liu, Q.; Zeng, W. Applications of transformer-based language models in bioinformatics: A survey. Bioinforma. Adv. 2023, 3, vbad001. doi: 10.1093/bioadv/vbad001
- Chi, J.; Shu, J.; Li, M.; Mudappathi, R.; Jin, Y.; Lewis, F.; Boon, A.; Qin, X.; Liu, L.; Gu, H. Artificial intelligence in metabolomics: A current review. TrAC Trends Anal. Chem. 2024, 178, 117852. doi: 10.1016/j.trac.2024.117852
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019.
- Lin, T.; Wang, Y.; Liu, X.; Qiu, X. A Survey of Transformers. AI Open 2021, 3, 111–132. doi: 10.1016/j.aiopen.2022.10.001
- Rashid, M.M.; Atilgan, N.; Dobres, J.; Day, S.; Penkova, V.; Küçük, M.; Steven; Clapp, R.; Sawyer, B.D. Humanizing AI in Education: A Readability Comparison of LLM and Human-Created Educational Content. Proc. Hum. Factors Ergon. Soc. Annu. Meet. 2024, 68, 596–603. https://doi.org/10.1177/10711813241261689.
- Naveed, H.; Khan, A.U.; Qiu, S.; Saqib, M.; Anwar, S.; Usman, M.; Akhtar, N.; Barnes, N.; Mian, A. A Comprehensive Overview of Large Language Models. arXiv 2024, arXiv:2307.06435.
- Radford, A.; Wu, J.; Child, R.; Luan, D.; Amodei, D.; Sutskever, I. Language Models are Unsupervised Multitask Learners. OpenAI Blog 2019, 1, 9.
- Elman, J.L. Finding structure in time. Cogn. Sci. 1990, 14, 179–211. doi: 10.1016/0364-0213(90)90002-E
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. doi: 10.1162/neco.1997.9.8.1735
- Chung, J.; Gulcehre, C.; Cho, K.; Bengio, Y. Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling. arXiv 2014, arXiv:1412.3555.
- O’Shea, K.; Nash, R. An Introduction to Convolutional Neural Networks. arXiv 2015, arXiv:1511.08458.
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to Sequence Learning with Neural Networks. Adv. Neural Inf. Process. Syst. 2014, 27.
- Li, M.; Wang, X.R. Peak alignment of gas chromatography–mass spectrometry data with deep learning. J. Chromatogr. A 2019, 1604, 460476. doi: 10.1016/j.chroma.2019.460476
- Seddiki, K.; Precioso, F.; Sanabria, M.; Salzet, M.; Fournier, I.; Droit, A. Early Diagnosis: End-to-End CNN–LSTM Models for Mass Spectrometry Data Classification. Anal. Chem. 2023, 95, 13431–13437. doi: 10.1021/acs.analchem.3c00613
- Jain, S.; Safo, S.E. DeepIDA-GRU: A deep learning pipeline for integrative discriminant analysis of cross-sectional and longitudinal multiview data with applications to inflammatory bowel disease classification. Brief. Bioinform. 2024, 25, bbae339. doi: 10.1093/bib/bbae339
- Kim, H.W.; Zhang, C.; Cottrell, G.W.; Gerwick, W.H. SMART-Miner: A convolutional neural network-based metabolite identification from 1H-13C HSQC spectra. Magn. Reson. Chem. 2022, 60, 1070–1075. doi: 10.1002/mrc.5240
- Tang, D.; Qin, B.; Liu, T. Document Modeling with Gated Recurrent Neural Network for Sentiment Classification. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Lisbon, Portugal, 17–21 September 2015; Association for Computational Linguistics: Lisbon, Portugal, 2015; pp. 1422–1432. https://doi.org/10.18653/v1/D15-1167.
- Huang, X.; Tan, H.; Lin, G.; Tian, Y. A LSTM-based bidirectional translation model for optimizing rare words and terminologies. In Proceedings of the 2018 International Conference on Artificial Intelligence and Big Data (ICAIBD), Chengdu, China, 26–28 May 2018; pp. 185–189. https://doi.org/10.1109/ICAIBD.2018.8396191.
- Radford, A.; Narasimhan, K. Improving Language Understanding by Generative Pre-Training. OpenAI 2018, 12.
- Raffel, C.; Shazeer, N.; Roberts, A.; Lee, K.; Narang, S.; Matena, M.; Zhou, Y.; Li, W.; Liu, P.J. Exploring the Limits of Transfer Learning with a Unified Text-to-Text Transformer. J. Mach. Learn. Res. 2020, 21, 1–67.
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. In Proceedings of the Ninth International Conference on Learning Representations, ICLR 2021, Virtual, 3–7 May 2021.
- Verma, P.; Berger, J. Audio Transformers:Transformer Architectures For Large Scale Audio Understanding. Adieu Convolutions. arXiv 2021, arXiv:2105.00335.
- Ma, Y.; Chi, D.; Wu, S.; Liu, Y.; Zhuang, Y.; Hao, J.; King, I. Actra: Optimized Transformer Architecture for Vision-Language-Action Models in Robot Learning. arXiv 2024, arXiv:2408.01147.
- Wang, T.; Roberts, A.; Hesslow, D.; Le Scao, T.; Chung, H.W.; Beltagy, I.; Launay, J.; Raffel, C. What Language Model Architecture and Pretraining Objective Work Best for Zero-Shot Generalization? In Proceedings of the 39th International Conference on Machine Learning 2022, Baltimore, MD, USA on 17–23 July 2022.
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901.
- Xue, L.; Constant, N.; Roberts, A.; Kale, M.; Al-Rfou, R.; Siddhant, A.; Barua, A.; Raffel, C. mT5: A massively multilingual pre-trained text-to-text transformer. arXiv 2020, arXiv:2010.11934. doi: 10.18653/v1/2021.naacl-main.41
- Chung, H.W.; Hou, L.; Longpre, S.; Zoph, B.; Tay, Y.; Fedus, W.; Li, Y.; Wang, X.; Dehghani, M.; Brahma, S.; et al. Scaling Instruction-Finetuned Language Models. J. Mach. Learn. Res. 2022, 25, 1–53.
- Ouyang, L.; Wu, J.; Jiang, X.; Almeida, D.; Wainwright, C.; Mishkin, P.; Zhang, C.; Agarwal, S.; Slama, K.; Ray, A.; et al. Training language models to follow instructions with human feedback. Adv. Neural Inf. Process. Syst. 2022, 35, 27730–27744.
- Xiao, Y.; Sun, E.; Jin, Y.; Wang, Q.; Wang, W. ProteinGPT: Multimodal LLM for Protein Property Prediction and Structure Understanding. arXiv 2024, arXiv:2408.11363.
- Wang, C.; Fan, H.; Quan, R.; Yang, Y. ProtChatGPT: Towards Understanding Proteins with Large Language Models. arXiv 2024, arXiv:2402.09649.
- Jin, Q.; Yang, Y.; Chen, Q.; Lu, Z. GeneGPT: Augmenting large language models with domain tools for improved access to biomedical information. Bioinformatics 2024, 40, btae075. doi: 10.1093/bioinformatics/btae075
- Yang, S.; Xu, P. LLM4THP: A computing tool to identify tumor homing peptides by molecular and sequence representation of large language model based on two-layer ensemble model strategy. Amino Acids 2024, 56, 62. doi: 10.1007/s00726-024-03422-5
- Liu, H.; Wang, H. GenoTEX: A Benchmark for Evaluating LLM-Based Exploration of Gene Expression Data in Alignment with Bioinformaticians. arXiv 2024, arXiv:2406.15341.
- Lin, X.; Deng, G.; Li, Y.; Ge, J.; Ho JW, K.; Liu, Y. GeneRAG: Enhancing Large Language Models with Gene-Related Task by Retrieval-Augmented Generation. bioRxiv 2024. https://doi.org/10.1101/2024.06.24.600176.
- Gao, Z.; Liu, Q.; Zeng, W.; Jiang, R.; Wong, W.H. EpiGePT: A pretrained transformer-based language model for context-specific human epigenomics. Genome Biol. 2024, 25, 310. doi: 10.1186/s13059-024-03449-7
- Chakraborty, C.; Bhattacharya, M.; Lee, S.-S. Artificial intelligence enabled ChatGPT and large language models in drug target discovery, drug discovery, and development. Mol. Ther. Nucleic Acids 2023, 33, 866–868. doi: 10.1016/j.omtn.2023.08.009
- Ma, T.; Lin, X.; Li, T.; Li, C.; Chen, L.; Zhou, P.; Cai, X.; Yang, X.; Zeng, D.; Cao, D.; et al. Y-Mol: A Multiscale Biomedical Knowledge-Guided Large Language Model for Drug Development. arXiv 2024, arXiv:2410.11550.
- Sheikholeslami, M.; Mazrouei, N.; Gheisari, Y.; Fasihi, A.; Irajpour, M.; Motahharynia, A. DrugGen: Advancing Drug Discovery with Large Language Models and Reinforcement Learning Feedback. arXiv 2024, arXiv:2411.14157. doi: 10.1038/s41598-025-98629-1
- Hu, M.; Alkhairy, S.; Lee, I.; Pillich, R.T.; Fong, D.; Smith, K.; Bachelder, R.; Ideker, T. Evaluation of large language models for discovery of gene set function. Nat. Methods 2024, 22, 82–91. https://doi.org/10.1038/s41592-024-02525-x.
- Liu, Y.; Chen, Z.; Wang, Y.G.; Shen, Y. TourSynbio-Search: A Large Language Model Driven Agent Framework for Unified Search Method for Protein Engineering. In Proceedings of the 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Lisbon, Portugal, 3–6 December 2024. doi: 10.1109/BIBM62325.2024.10822318
- Van Kempen, M.; Kim, S.S.; Tumescheit, C.; Mirdita, M.; Lee, J.; Gilchrist, C.L.M.; Söding, J.; Steinegger, M. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 2024, 42, 243–246. doi: 10.1038/s41587-023-01773-0
- Bran, A.M.; Cox, S.; Schilter, O.; Baldassari, C.; White, A.D.; Schwaller, P. Augmenting large language models with chemistry tools. Nat. Mach. Intell. 2024, 6, 525–535. doi: 10.1038/s42256-024-00832-8
- Boiko, D.A.; MacKnight, R.; Kline, B.; Gomes, G. Autonomous chemical research with large language models. Nature 2023, 624, 570–578. doi: 10.1038/s41586-023-06792-0
- Yang, E.-W.; Velazquez-Villarreal, E. AI-driven conversational agent enhances clinical and genomic data integration for precision medicine research. medRxiv 2024. https://doi.org/10.1101/2024.11.27.24318113.
- Chen, Q.; Deng, C. Bioinfo-Bench: A Simple Benchmark Framework for LLM Bioinformatics Skills Evaluation. bioRxiv 2023. https://doi.org/10.1101/2023.10.18.563023.
- Zhou, J.; Zhang, B.; Chen, X.; Li, H.; Xu, X.; Chen, S.; Gao, X. Automated Bioinformatics Analysis via AutoBA. arXiv 2023, arXiv:2309.03242.
- Biana, J.; Zhai, W.; Huang, X.; Zheng, J.; Zhu, S. VANER: Leveraging Large Language Model for Versatile and Adaptive Biomedical Named Entity Recognition. arXiv 2024, arXiv:2404.17835. doi: 10.3233/FAIA240664
- Bian, J.; Zheng, J.; Zhang, Y.; Zhou, H.; Zhu, S. One-shot Biomedical Named Entity Recognition via Knowledge-Inspired Large Language Model. In Proceedings of the 15th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Shenzhen China, 22–25 November 2024; Association for Computing Machinery: New York, NY, USA, 2024. https://doi.org/10.1145/3698587.3701356.
- Wei, J.; Zhuo, L.; Fu, X.; Zeng, X.; Wang, L.; Zou, Q.; Cao, D. DrugReAlign: A multisource prompt framework for drug repurposing based on large language models. BMC Biol. 2024, 22, 226. doi: 10.1186/s12915-024-02028-3
- Li, Y.; Gao, C.; Song, X.; Wang, X.; Xu, Y.; Han, S. DrugGPT: A GPT-based Strategy for Designing Potential Ligands Targeting Specific Proteins. bioRxiv 2023. https://doi.org/10.1101/2023.06.29.543848.
- Liu, Y.; Ding, S.; Zhou, S.; Fan, W.; Tan, Q. MolecularGPT: Open Large Language Model (LLM) for Few-Shot Molecular Property Prediction. arXiv 2024, arXiv:2406.12950.
- Zhang, D.; Liu, W.; Tan, Q.; Chen, J.; Yan, H.; Yan, Y.; Li, J.; Huang, W.; Yue, X.; Ouyang, W.; et al. ChemLLM: A Chemical Large Language Model. arXiv 2024, arXiv:2402.06852.
- Galkin, F.; Naumov, V.; Pushkov, S.; Sidorenko, D.; Urban, A.; Zagirova, D.; Alawi, K.M.; Aliper, A.; Gumerov, R.; Kalashnikov, A.; et al. Precious3GPT: Multimodal Multi-Species Multi-Omics Multi-Tissue Transformer for Aging Research and Drug Discovery. bioRxiv 2024. https://doi.org/10.1101/2024.07.25.605062.
- Elsborg, J.; Salvatore, M. Using LLMs and Explainable ML to Analyze Biomarkers at Single-Cell Level for Improved Understanding of Diseases. Biomolecules 2023, 13, 1516. doi: 10.3390/biom13101516
- Baygi, S.F.; Barupal, D.K. IDSL_MINT: A deep learning framework to predict molecular fingerprints from mass spectra. J. Cheminform. 2024, 16, 8. doi: 10.1186/s13321-024-00804-5
- Würf, V.; Köhler, N.; Molnar, F.; Hahnefeld, L.; Gurke, R.; Witting, M.; Pauling, J.K. LipiDetective—A deep learning model for the identification of molecular lipid species in tandem mass spectra. bioRxiv 2024. https://doi.org/10.1101/2024.10.07.617094 2024.
- Butler, T.; Frandsen, A.; Lightheart, R.; Bargh, B.; Taylor, J.; Bollerman, T.J.; Kerby, T.; West, K.; Voronov, G.; Moon, K.; et al. MS2Mol: A transformer model for illuminating dark chemical space from mass spectra. ChemRxiv 2023. https://doi.org/10.26434/chemrxiv-2023-vsmpx-v3 2023.
- Xue, J.; Wang, B.; Ji, H.; Li, W. RT-Transformer: Retention time prediction for metabolite annotation to assist in metabolite identification. Bioinformatics 2024, 40, btae084. doi: 10.1093/bioinformatics/btae084
- Young, A.; Wang, B.; Röst, H. MassFormer: Tandem Mass Spectrum Prediction for Small Molecules using Graph Transformers. Nat. Mach. Intell. 2023, 6, 404–416.
- Liu, Y.; Yoshizawa, A.C.; Ling, Y.; Okuda, S. Insights into predicting small molecule retention times in liquid chromatography using deep learning. J. Cheminformatics 2024, 16, 113. doi: 10.1186/s13321-024-00905-1
- Cao, M.; Fraser, K.; Huege, J.; Featonby, T.; Rasmussen, S.; Jones, C. Predicting retention time in hydrophilic interaction liquid chromatography mass spectrometry and its use for peak annotation in metabolomics. Metabolomics 2015, 11, 696–706. doi: 10.1007/s11306-014-0727-x
- Yang, Q.; Ji, H.; Lu, H.; Zhang, Z. Prediction of Liquid Chromatographic Retention Time with Graph Neural Networks to Assist in Small Molecule Identification. Anal. Chem. 2021, 93, 2200–2206. doi: 10.1021/acs.analchem.0c04071
- Osipenko, S.; Nikolaev, E.; Kostyukevich, Y. Retention Time Prediction with Message-Passing Neural Networks. Separations 2022, 9, 291. doi: 10.3390/separations9100291
- Ju, R.; Liu, X.; Zheng, F.; Lu, X.; Xu, G.; Lin, X. Deep Neural Network Pretrained by Weighted Autoencoders and Transfer Learning for Retention Time Prediction of Small Molecules. Anal. Chem. 2021, 93, 15651–15658. doi: 10.1021/acs.analchem.1c03250
- Fedorova, E.S.; Matyushin, D.D.; Plyushchenko, I.V.; Stavrianidi, A.N.; Buryak, A.K. Deep learning for retention time prediction in reversed-phase liquid chromatography. J. Chromatogr. A 2022, 1664, 462792. doi: 10.1016/j.chroma.2021.462792
- Zhao, S.; Li, L. Chemical derivatization in LC-MS-based metabolomics study. TrAC Trends Anal. Chem. 2020, 131, 115988. doi: 10.1016/j.trac.2020.115988
- Domingo-Almenara, X.; Montenegro-Burke, J.R.; Benton, H.P.; Siuzdak, G. Annotation: A Computational Solution for Streamlining Metabolomics Analysis. Anal. Chem. 2018, 90, 480–489. doi: 10.1021/acs.analchem.7b03929
- Lenski, M.; Maallem, S.; Zarcone, G.; Garçon, G.; Lo-Guidice, J.M.; Anthérieu, S.; Allorge, D. Prediction of a Large-Scale Database of Collision Cross-Section and Retention Time Using Machine Learning to Reduce False Positive Annotations in Untargeted Metabolomics. Metabolites 2023, 13, 282. doi: 10.3390/metabo13020282
- Voronov, G.; Frandsen, A.; Bargh, B.; Healey, D.; Lightheart, R.; Kind, T.; Dorrestein, P.; Colluru, V.; Butler, T. MS2Prop: A machine learning model that directly predicts chemical properties from mass spectrometry data for novel compounds. bioRxiv 2022. https://doi.org/10.1101/2022.10.09.511482.
- Heiles, S. Advanced tandem mass spectrometry in metabolomics and lipidomics—Methods and applications. Anal. Bioanal. Chem. 2021, 413, 5927–5948. doi: 10.1007/s00216-021-03425-1
- Chen, B.; Li, H.; Huang, R.; Tang, Y.; Li, F. Deep learning prediction of electrospray ionization tandem mass spectra of chemically derived molecules. Nat. Commun. 2024, 15, 8396. doi: 10.1038/s41467-024-52805-5
- Ekvall, M.; Truong, P.; Gabriel, W.; Wilhelm, M.; Käll, L. Prosit Transformer: A transformer for Prediction of MS2 Spectrum Intensities. J. Proteome Res. 2022, 21, 1359–1364. doi: 10.1021/acs.jproteome.1c00870
- Young, A.; Röst, H.; Wang, B. Tandem mass spectrum prediction for small molecules using graph transformers. Nat. Mach. Intell. 2024, 6, 404–416. doi: 10.1038/s42256-024-00816-8
- Allen, F.; Greiner, R.; Wishart, D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 2015, 11, 98–110. doi: 10.1007/s11306-014-0676-4
- Wang, F.; Liigand, J.; Tian, S.; Arndt, D.; Greiner, R.; Wishart, D.S. CFM-ID 4.0: More Accurate ESI-MS/MS Spectral Prediction and Compound Identification. Anal. Chem. 2021, 93, 11692–11700. doi: 10.1021/acs.analchem.1c01465
- Wei, J.N.; Belanger, D.; Adams, R.P.; Sculley, D. Rapid Prediction of Electron–Ionization Mass Spectrometry Using Neural Networks. ACS Cent. Sci. 2019, 5, 700–708. doi: 10.1021/acscentsci.9b00085
- Jin, W.; Coley, C.W.; Barzilay, R.; Jaakkola, T. Predicting Organic Reaction Outcomes with Weisfeiler-Lehman Network. In Proceedings of the 2017 Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017.
- Wenk, M. R. Lipidomics: New Tools and Applications. Cell 2010, 143, 888–895. doi: 10.1016/j.cell.2010.11.033
- Kloska, A.; Węsierska, M.; Malinowska, M.; Gabig-Cimińska, M.; Jakóbkiewicz-Banecka, J. Lipophagy and Lipolysis Status in Lipid Storage and Lipid Metabolism Diseases. Int. J. Mol. Sci. 2020, 21, 6113. doi: 10.3390/ijms21176113
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structures and tools. Lipidomics Imaging Mass Spectrom. 2011, 1811, 637–647. doi: 10.1016/j.bbalip.2011.06.009
- Gerhardtova, I.; Jankech, T.; Majerova, P.; Piestansky, J.; Olesova, D.; Kovac, A.; Jampilek, J. Recent Analytical Methodologies in Lipid Analysis. Int. J. Mol. Sci. 2024, 25, 2249. doi: 10.3390/ijms25042249
- Yang, J.; Cai, Y.; Zhao, K.; Xie, H.; Chen, X. Concepts and applications of chemical fingerprint for hit and lead screening. Drug Discov. Today 2022, 27, 103356. doi: 10.1016/j.drudis.2022.103356
- Muegge, I.; Mukherjee, P. An overview of molecular fingerprint similarity search in virtual screening. Expert Opin. Drug Discov. 2015, 11, 137–148. doi: 10.1517/17460441.2016.1117070
- Cereto-Massagué; A; Ojeda, M.J.; Valls, C.; Mulero, M.; Garcia-Vallvé; S; Pujadas, G. Molecular fingerprint similarity search in virtual screening. Methods 2015, 71, 58–63. doi: 10.1016/j.ymeth.2014.08.005
- Goldman, S.; Wohlwend, J.; Stražar, M.; Haroush, G.; Xavier, R.J.; Coley, C.W. Annotating metabolite mass spectra with domain-inspired chemical formula transformers. Nat. Mach. Intell. 2023, 5, 965–979. doi: 10.1038/s42256-023-00708-3
- Stravs, M.A.; Dührkop, K.; Böcker, S.; Zamboni, N. MSNovelist: De novo structure generation from mass spectra. Nat. Methods 2022, 19, 865–870. doi: 10.1038/s41592-022-01486-3
- Litsa, E.E.; Chenthamarakshan, V.; Das, P.; Kavraki, L.E. An end-to-end deep learning framework for translating mass spectra to de-novo molecules. Commun. Chem. 2023, 6, 132. doi: 10.1038/s42004-023-00932-3
- Shrivastava, A.D.; Swainston, N.; Samanta, S.; Roberts, I.; Wright Muelas, M.; Kell, D.B. MassGenie: A Transformer-Based Deep Learning Method for Identifying Small Molecules from Their Mass Spectra. Biomolecules 2021, 11, 1793. doi: 10.3390/biom11121793
- Elser, D.; Huber, F.; Gaquerel, E. Mass2SMILES: Deep learning based fast prediction of structures and functional groups directly from high-resolution MS/MS spectra. bioRxiv 2023. https://doi.org/10.1101/2023.07.06.547963.
- Vaniya, A.; Fiehn, O. Using fragmentation trees and mass spectral trees for identifying unknown compounds in metabolomics. TrAC Trends Anal. Chem. 2015, 69, 52–61. doi: 10.1016/j.trac.2015.04.002
- Zhang, M.; Xia, Y.; Wu, N.; Qian, K.; Zeng, J. MS2-Transformer: An End-to-End Model for MS/MS-assisted Molecule Identification. 2022.
- Yang, Y.; Sun, S.; Yang, S.; Yang, Q.; Lu, X.; Wang, X.; Yu, Q.; Huo, X.; Qian, X. Structural annotation of unknown molecules in a miniaturized mass spectrometer based on a transformer enabled fragment tree method. Commun. Chem. 2024, 7, 109. doi: 10.1038/s42004-024-01189-0
- Meng, W.; Pan, H.; Sha, Y.; Zhai, X.; Xing, A.; Lingampelly, S.S.; Sripathi, S.R.; Wang, Y.; Li, K. Metabolic Connectome and Its Role in the Prediction, Diagnosis, and Treatment of Complex Diseases. Metabolites 2024, 14, 93. doi: 10.3390/metabo14020093
- Amara, A.; Frainay, C.; Jourdan, F.; Naake, T.; Neumann, S.; Novoa-Del-Toro, E.M.; Salek, R.M.; Salzer, L.; Scharfenberg, S.; Witting, M. Networks and Graphs Discovery in Metabolomics Data Analysis and Interpretation. Front. Mol. Biosci. 2022, 9, 841373. doi: 10.3389/fmolb.2022.841373
- Matsumoto, N.; Moran, J.; Choi, H.; Hernandez, M.E.; Venkatesan, M.; Wang, P.; Moore, J.H. KRAGEN: A knowledge graph-enhanced RAG framework for biomedical problem solving using large language models. Bioinformatics 2024, 40, btae353. doi: 10.1093/bioinformatics/btae353
- Wen, Y.; Wang, Z.; Sun, J. MindMap: Knowledge Graph Prompting Sparks Graph of Thoughts in Large Language Models. In Proceedings of the 62nd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Bangkok, Thailand, 11–16 August 2024; Association for Computational Linguistics, Bangkok, Thailand, 2024; pp. 10370–10388. https://doi.org/10.18653/v1/2024.acl-long.558.
- Beck, A.G.; Muhoberac, M.; Randolph, C.E.; Beveridge, C.H.; Wijewardhane, P.R.; Kenttamaa, H.I.; Chopra, G. Recent Developments in Machine Learning for Mass Spectrometry. ACS Meas. Sci. Au 2024, 4, 233–246. doi: 10.1021/acsmeasuresciau.3c00060
- Pinto, R.C.; Karaman, I.; Lewis, M.R.; Hällqvist, J.; Kaluarachchi, M.; Graça, G.; Chekmeneva, E.; Durainayagam, B.; Ghanbari, M.; Ikram, M.A.; et al. Finding Correspondence between Metabolomic Features in Untargeted Liquid Chromatography–Mass Spectrometry Metabolomics Datasets. Anal. Chem. 2022, 94, 5493–5503. doi: 10.1021/acs.analchem.1c03592
- Wörheide, M.A.; Krumsiek, J.; Kastenmüller, G.; Arnold, M. Multi-omics integration in biomedical research—A metabolomics-centric review. Anal. Chim. Acta 2021, 1141, 144–162. doi: 10.1016/j.aca.2020.10.038
- Sanches PH, G.; de Melo, N.C.; Porcari, A.M.; de Carvalho, L.M. Integrating Molecular Perspectives: Strategies for Comprehensive Multi-Omics Integrative Data Analysis and Machine Learning Applications in Transcriptomics, Proteomics, and Metabolomics. Biology 2024, 13, 848. doi: 10.3390/biology13110848
- Maan, K.; Baghel, R.; Dhariwal, S.; Sharma, A.; Bakhshi, R.; Rana, P. Metabolomics and transcriptomics based multi-omics integration reveals radiation-induced altered pathway networking and underlying mechanism. NPJ Syst. Biol. Appl. 2023, 9, 42. doi: 10.1038/s41540-023-00305-5





